BOKEI Bayesian Optimization Using Knowledge of Correlated Torsions and Expected Improvement for Conformer Generation
Leung Sing Chan, Geoffrey R. Hutchison, Garrett Morris. “BOKEI Bayesian Optimization Using Knowledge of Correlated Torsions and Expected Improvement for Conformer Generation” Phys. Chem. Chem. Phys. (2020) 22(9) 5211-5219. DOI Preprint
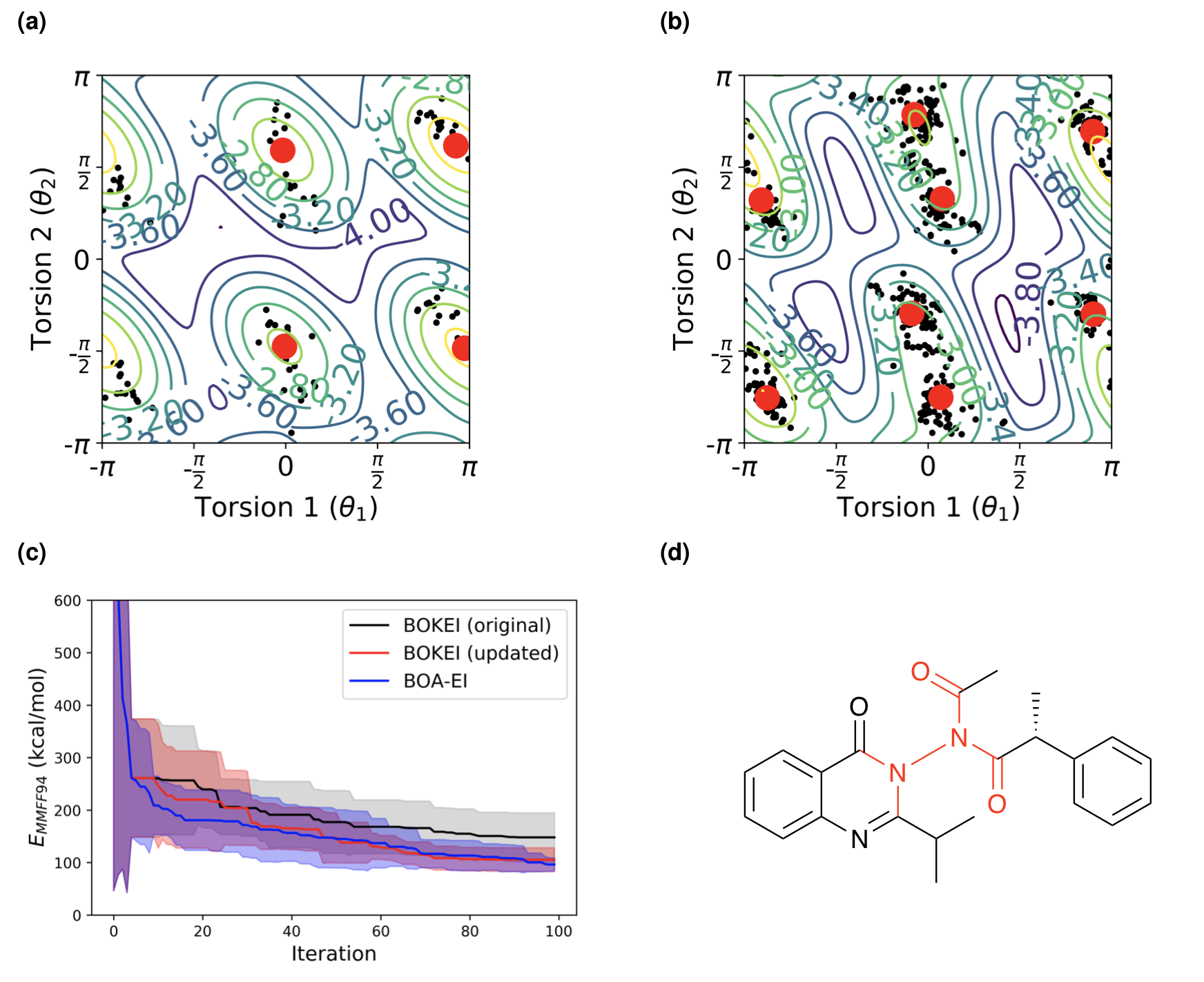 A key challenge in conformer sampling is to find low-energy conformations with a small number of energy evaluations. We have recently demonstrated Bayesian optimization as an effective method to search for energetically favorable conformations. This approach balances between exploitation and exploration, and lead to superior performance when compared to exhaustive or random search methods. In this work, we extend strategies on proteins and oligopeptides (eg Ramachandran plots of secondary structure) to study the correlated torsions in small molecules. We use a bivariate von Mises distribution to capture the correlations, and use it to constrain the search space. We validate the performance of our Bayesian optimization with prior knowledge (BOKEI) on a dataset consisting of 533 diverse small organic molecules, using a force field (MMFF94) and a semi-empirical method (GFN2). We compare BOKEI with Bayesian optimization with expected improvement (BOA-EI), and a genetic algorithm (GA), using a fixed number of energy evaluations. In 70 (±2.1)% of the cases examined, BOKEI finds lower energy conformations than global optimization with BOA-EI or GA. More importantly, these patterns find correlated torsions in 10-15% of molecules in larger data sets, 3-8 times more frequently than previous work. We also find that the BOKEI patterns not only describe steric clashes, but also reflect favorable intramolecular interactions, including hydrogen bonds and π-π stacking. Further understanding of the conformational preferences of molecules will help find low energy conformers efficiently for a wide range of computational modeling applications.
A key challenge in conformer sampling is to find low-energy conformations with a small number of energy evaluations. We have recently demonstrated Bayesian optimization as an effective method to search for energetically favorable conformations. This approach balances between exploitation and exploration, and lead to superior performance when compared to exhaustive or random search methods. In this work, we extend strategies on proteins and oligopeptides (eg Ramachandran plots of secondary structure) to study the correlated torsions in small molecules. We use a bivariate von Mises distribution to capture the correlations, and use it to constrain the search space. We validate the performance of our Bayesian optimization with prior knowledge (BOKEI) on a dataset consisting of 533 diverse small organic molecules, using a force field (MMFF94) and a semi-empirical method (GFN2). We compare BOKEI with Bayesian optimization with expected improvement (BOA-EI), and a genetic algorithm (GA), using a fixed number of energy evaluations. In 70 (±2.1)% of the cases examined, BOKEI finds lower energy conformations than global optimization with BOA-EI or GA. More importantly, these patterns find correlated torsions in 10-15% of molecules in larger data sets, 3-8 times more frequently than previous work. We also find that the BOKEI patterns not only describe steric clashes, but also reflect favorable intramolecular interactions, including hydrogen bonds and π-π stacking. Further understanding of the conformational preferences of molecules will help find low energy conformers efficiently for a wide range of computational modeling applications.